New Genetic Tools Have Dramatically Changed Wildlife Conservation
On a sunny fall morning, biologist Andy Hubbard set up a makeshift lab next to small pools at a national park’s outcropping of ancient granite rocks. Immersed in the stillness of a cactus forest, he and his team filled tiny bags with murky water and meticulously strained it through miniscule filters.

Those filters would later be sent to a laboratory to test for genetic material shed by animals in the water. By collecting environmental DNA, or eDNA, Hubbard and three other members of a National Park Service team hoped to detect signs of native critters and the invasive bullfrogs that have proved devastating for their existence.
“This technology will end up being critical because it’s a more efficient way to detect invasive species, as well as rare species,” said Hubbard, program manager for the National Park Service Sonoran Desert Network in Tucson, Arizona.
Around the world, scientists like Hubbard are increasingly turning to eDNA to detect species from discarded bits of skin, scales, and mucus in water, soil, and air. In the field of conservation research, the emerging technology is opening new frontiers to monitor endangered species, track invasive ones, and sample general biodiversity. It’s also cheaper. And while the field still faces limitations around accuracy and precision, scientists say eDNA is fast becoming a game-changer for wildlife conservation efforts.
“Environmental DNA has become more and more important as we are putting increased emphasis on understanding biodiversity and importantly, biodiversity loss,” said Adam Sepulveda, a research scientist with the U.S. Geological Survey’s Northern Rocky Mountain Science Center in Bozeman, Montana.

In such a rapidly growing field, technological innovation has been key to advancing eDNA methodologies, especially as scientists attempt to move past constraints in the field: While some researchers continue to use traditional tools, like visual surveys, traps, and nets, others are banking on portable robots to collect more frequent samples, or advanced lab analysis methods to take stock of biodiversity. Meanwhile, Sepulveda and other scientists have said there should be a national strategy for eDNA application, which, they say, would avoid inconsistent guidelines across agencies, and make the eDNA analysis more efficient.
The scientific community now recognizes that eDNA is a useful lens through which much can be learned about species that are difficult to identify using only traditional tools, said Sepulveda: “eDNA gives us some power to understand what could be happening with these harder-to-find species.”
The exploration of environmental DNA started in the 1980s with researchers studying microbes or microorganisms, such as bacteria, fungi, and algae. But it wasn’t until the early 2000s that the use of eDNA started gaining momentum as methods and technologies evolved, and more scientists began adopting the approach. Research into aquatic ecosystems exploded after the 2008 publication of a study on eDNA testing from samples collected in wetlands in France that detected the presence of invasive American bullfrogs.
That arena of early detection and management of invasive species has been one where eDNA has held particular promise. Rather than conduct time-intensive, often difficult and intrusive surveys, researchers can now rely on a tiny sample of eDNA to determine the existence of species before they cause serious harm to ecosystems, as well as organisms too small to see with the naked eye.
The technology is a particularly useful tool for tracking invasive species because of its high sensitivity. It “can detect DNA evidence from just a few individuals at the start of an invasion,” Sepulveda wrote in an email.
Time is key, since once invasive species spread and get established in a new environment, they pose a major threat to native animals and their habitats. They can also cause significant destruction to infrastructure. For example, the diminutive zebra mussel, a shellfish discovered in the Great Lakes in the 1980s, can clog water intake for power and water plants and damage boats and equipment. It’s believed that zebra mussels first arrived with ballast water discharged from European ships, and they now live in the Mississippi River Basin, Great Lakes, and many other bodies of water throughout the nation.
“This technology will end up being critical because it’s a more efficient way to detect invasive species, as well as rare species.”
Such unfettered expansion comes at a high price: The annual cost of managing invasive species in the United States soared from $2 billion per year in the 1960s to $21 billion per year since 2010, according to a study published online in 2021. Globally, the economic cost related to invasive species over nearly 50 years is estimated at around $1.3 trillion.
In southern Arizona, the American bullfrog — which is native to the eastern U.S. and Canada — is a major source of distress. The voracious critters eat everything in their path, destroying native species like the federally threatened Chiricahua leopard frog.
Not only do the amphibians compete with smaller native species of frogs for food and space, but they also can spread disease-causing pathogens like chytrid fungus and ranaviruses, which in turn also contribute to native populations’ decline. “The American bullfrog is probably the most important and impactful non-native invasive animal in the Southwest,” Hubbard said.
In the hopes of tracking their spread, Hubbard and his springs-monitoring crew have collected eDNA in several national parks within Arizona and neighboring New Mexico.
At the eDNA collection last fall, Hubbard’s team spent an entire morning at Tucson’s Saguaro National Park, scooping out and filtering 1,000 milliliters — roughly four cups — of water from the pool. Filters the size of a dollar coin trap loose cells and DNA present in water samples, so team members carefully folded them with tweezers into tiny triangles and placed them in sterilized test tubes. Hubbard expects to have lab results from the DNA analysis by the early part of the year.



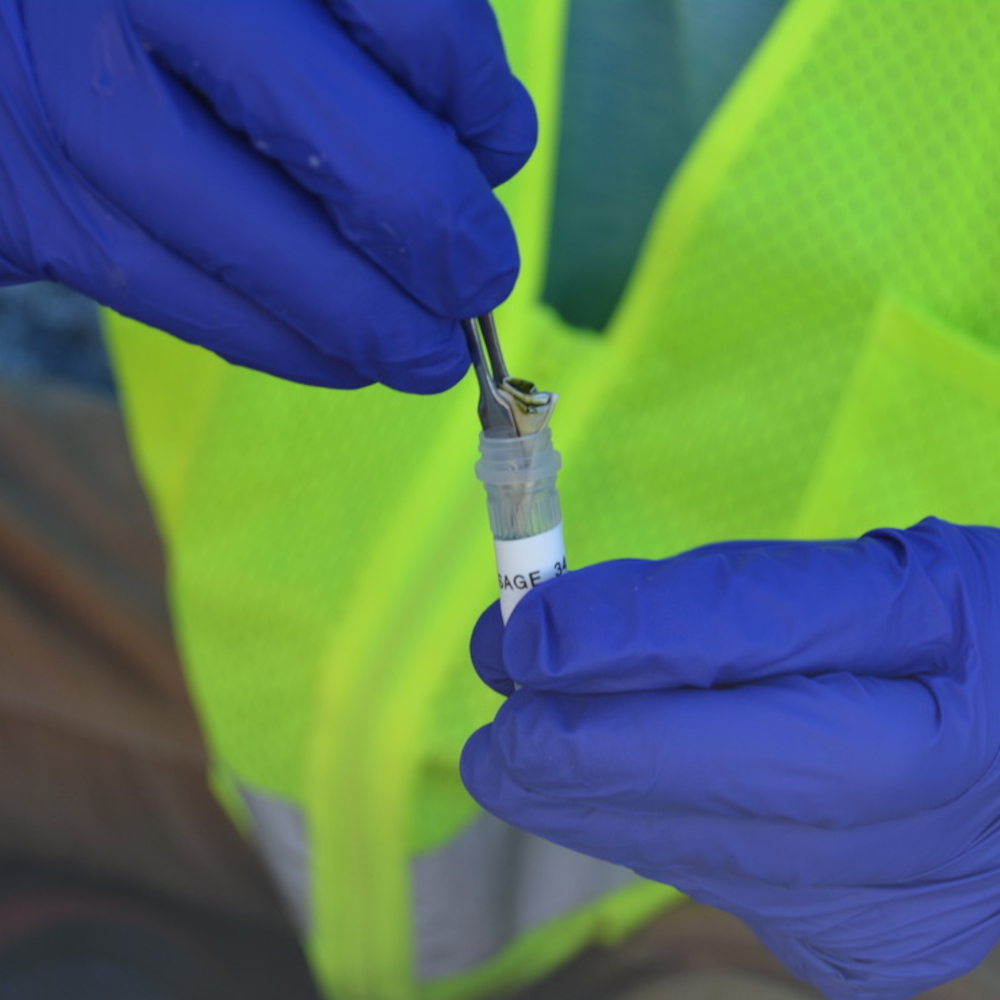

Like other researchers, Hubbard has grappled with some of eDNA’s limitations. For one, DNA can degrade quickly in water, particularly in warm weather. “As the water temperature increases, the remaining DNA starts to break down,” he said.
And eDNA does not shed light on species abundance, such as that of declining native frog species. A spring called “The Grotto” in Saguaro National Park “has lowland leopard frogs based on my results, but I can’t tell you how many, and I can’t tell you if that number has changed over time,” he said. The technology can only tell whether certain eDNA is present or not.
He’s also learned that eDNA may not be suitable for detection of certain species, such as those that shed sparse genetic material. That includes the threatened northern Mexican garter snake, which visual surveys previously have recorded in marshy areas, but eDNA has yet to uncover.
“We’re trying to optimize our sampling for multiple species,” Hubbard said. “And there’s always a trade-off. When you do that, there are going to be different approaches that will be more or less successful in detecting species, depending on their habitat or on their behaviors. So, this probably isn’t the best way to detect northern Mexican garter snakes.”
“Environmental DNA has become more and more important as we are putting increased emphasis on understanding biodiversity and importantly, biodiversity loss.”
Hubbard said the possibility of false negatives, when eDNA can miss species that may be present in the environment, or false positives, when the technology can detect species that are absent, could also pose challenges. Nevertheless, in the case of the garter snake, he said potential errors may be overcome with intensified water sampling combined with traditional tracking methods, such as placing wooden boards for snakes to hide under in wetland habitats. This allows researchers to lure snakes into areas they can easily access during surveys.
Where Hubbard sees the most value for the eDNA parks project is for monitoring frog populations to set conservation actions for native species, which include the eradication of invasive bullfrogs detected. “That’s our highest priority and highest concern right now,” he said.
Another limitation of the technology is related to sampling: While Hubbard’s team spent a few hours filtering water, they couldn’t stay all day — or overnight. Sepulveda, the USGS research scientist, says that robots roughly the size of carry-on luggage could help.
At the Northern Rocky Mountain Science Center, he and his colleagues are working on how best to incorporate eDNA and robot technology to address shortcomings in freshwater bodies battling the infestation of zebra mussels, quagga mussels — originally from Eastern Europe — and other aquatic invasive species. By using robots that can collect multiple eDNA samples over longer periods, fewer researchers will have to do the task.
Sepulveda’s work largely focuses on invasive species and supporting natural resource managers throughout the country in dealing with them, including those in the Columbia River Basin. “That is — or was until a few months ago — the last major water basin in the lower 48 to not yet be invaded by zebra or quagga mussels,” he said.
In September, quagga mussels were found in Idaho’s Snake River, the largest tributary of the Columbia River. The Columbia flows through seven states and a Canadian province. Natural resources agencies continue to monitor the area after moving quickly to contain the invasive species with a chemical treatment that also killed thousands of fish. Trying to control mussel infestation is an uphill battle, Sepulveda said. Female zebra and quagga mussels can produce up to 1 million eggs a year. Besides destroying native aquatic life, mussels attach themselves to hard surfaces like boat hulls that can carry them over long distances.
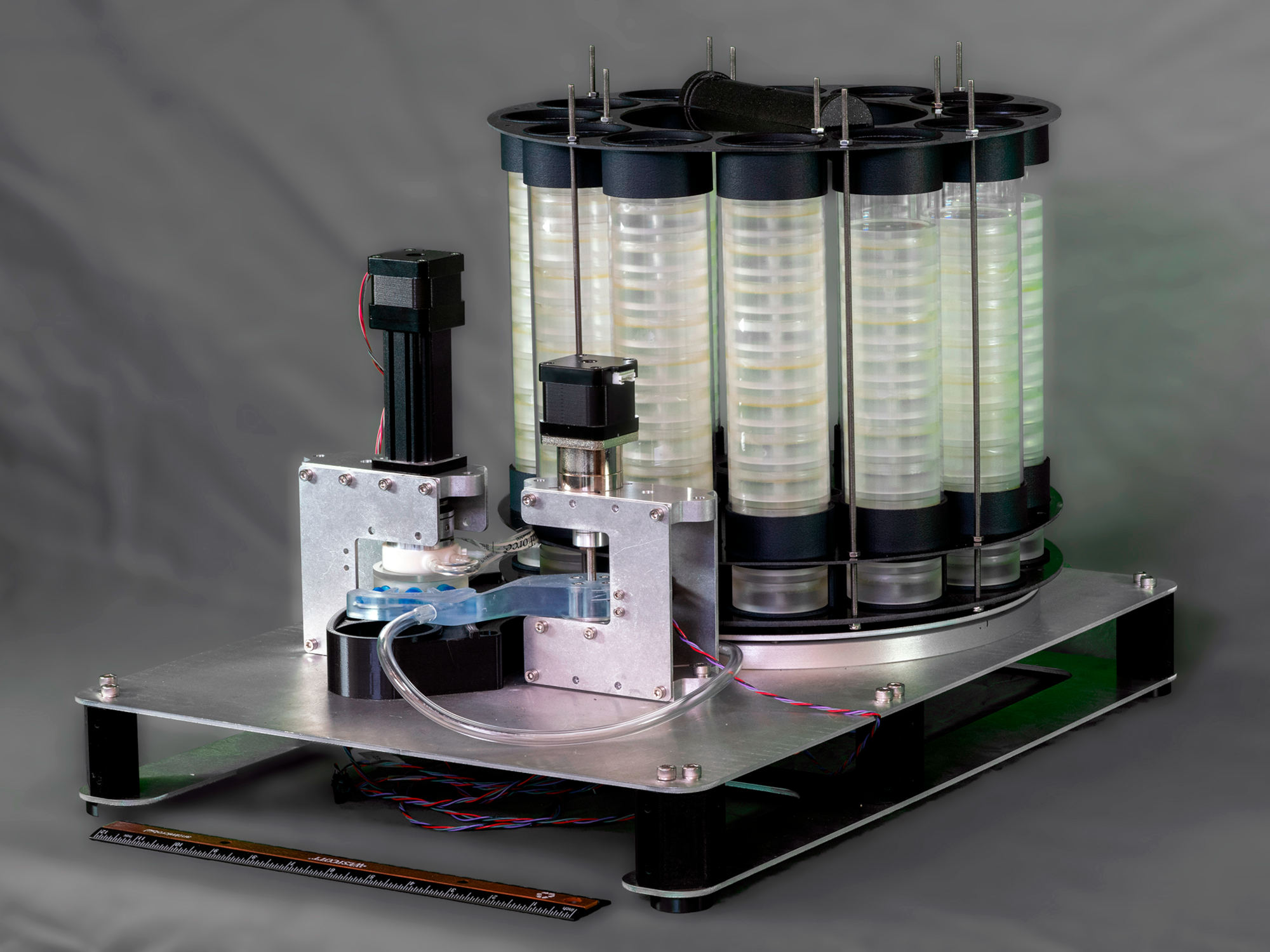
The researchers have tested robots that can collect eDNA water samples to reduce the cost of monitoring for invasive species and overcome some of the pitfalls of the technology — like the false negative results of the northern Mexican garter snake known to slither in the wild.
The robots, also called autonomous samplers, are designed to filter and preserve eDNA not only from invasive species, but also any aquatic and semi-aquatic organisms that leave genetic traces behind. Sepulveda leads a project dubbed READI-Net — short for “Rapid environmental DNA Assessment and Deployment Initiative and Network” — in which researchers are developing a leaner version of a robot that the Monterey Bay Aquarium Research Institute in Moss Landing, California, built some years ago for ocean exploration.



When ready for use in freshwater, the robots will be set up on shores, on fixed floating docks, or on boats. “They’re not meant to be submerged,” Sepulveda said of the robots. “They have tubes that go into the water to pull the water in, but the actual robotics are meant to be on land.”
Unlike humans, the freshwater robots will be able to collect samples much more frequently, day or night, increasing the likelihood of species detection. Collecting eDNA can be a challenge, said Sepulveda, because the currents that form in lakes, rivers, and oceans can carry it away from the organisms that discard it. And given its continued decay, the more time that passes, the harder it is to capture it.
“If you get your scoop of water, there is a chance, especially when something is very rare, that you were there 10 minutes too early or potentially 10 minutes too late — or a day too early, or a day too late,” he said. “And so, one of the ways that we can increase our ability to detect a new invader, just like our ability to detect the Covid virus, is by collecting more samples.”
The robots will be able to collect up to 144 samples before a scientist or technician would need to gather them for analysis, he said. “It can be left to collect samples for weeks to months.”
“If you get your scoop of water, there is a chance, especially when something is very rare, that you were there 10 minutes too early or potentially 10 minutes too late — or a day too early, or a day too late.”
Jim Birch, director of the SURF Center — Sensors: Underwater Research of the Future — at the Monterey Bay Aquarium Research Institute, likened the robots to a stripped-down version of the environmental sample processor, an older underwater robot originally built to study harmful algal blooms, also called red tides, that can kill animals.
As eDNA technology became more popular, Birch said, two postdoctoral researchers at the institute tested whether the robot could successfully collect eDNA samples from the aquarium. The experiment worked. Filtered water samples not only picked DNA traces of fish, but also detected genetic bits of the turkey and chicken feed that fish had gobbled up.
After some field testing, the robot is now being redesigned for optimal use in freshwater, Birch said: “It will have a big effect on invasive species, but its use is much broader.”
Sepulveda said field deployments of a prototype are just beginning this year. “We’re hoping by 2025 to have 10 that are ready to go and that we can get out to our partners,” he said.
The robotic eDNA-gathering technique will be especially useful in programs with limited manpower, he said. “If we can detect these invasive species and figure out what they are and where they are very early in the invasion process, then we’re likely to limit the economic and ecological costs of that invasion.”

The collection of eDNA is just the first step in trying to identify species from the bits of themselves they leave behind as they roam different habitats. The dead skin, saliva, scat, and other cellular material that organisms shed must then be analyzed in a laboratory using molecular methods.
At Washington State University’s School of the Environment, associate professor Caren Goldberg extracts the DNA trapped in the filters that Hubbard sends from southern Arizona. “We do one species at a time, and then sometimes we have to do some extra cleaning on the samples, and then we process all the data, and we double-check it to make sure everything looks good before sending it back,” she said.
Environmental technology is a valuable tool for finding elusive species like frogs, Goldberg said. She knows how slippery the creatures can be because, as a University of Arizona student, she completed her master’s thesis after chasing barking frogs in the mountains and surveying Chiricahua leopard frogs that can be difficult to see in murky, and often deep, water holes.
“If we can detect these invasive species and figure out what they are and where they are very early in the invasion process, then we’re likely to limit the economic and ecological costs of that invasion.”
“The reason why I could see how exciting the whole field would be is because I spent so much time out in Arizona looking at these ponds and not being able to find the frogs,” she said.
Goldberg now works with various federal and state agencies using eDNA techniques in their conservation programs. For the southern Arizona project, she looks for genetic signs of such species as local leopard frogs and the large bullfrogs that devour the small, native amphibians, contributing to their population drop. The federal government considers the Chiricahua leopard frog, which now emits its snore-like mating call in a shrinking habitat, a threatened species.
In earlier samples from Hubbard, Goldberg has detected the invasive frog. She has also tested for the presence of endangered jaguars that trail cameras have photographed in the borderlands. The sample was from ponds where Hubbard had hoped a stealthy, thirsty feline might have left its saliva, but no DNA traces were found.


Since Goldberg seeks to detect single species at a time, she uses a laboratory technique called quantitative polymerase chain reaction, or qPCR. It’s an analytical technique similar to the one used to test for Covid-19 infection. And while the methodology is widely used in single-species detection, it’s not suitable to detect multiple species from a mixed sample. Instead, scientists are increasingly using eDNA metabarcoding, which is a relatively new approach that can detect a large number of DNA sequences.
The advent of eDNA metabarcoding and next generation sequencing has been a breakthrough, said Jan Gogarten, an evolutionary community ecologist at the Helmholtz Institute for One Health in Germany. “We can sequence the diversity in a single sample, and that unlocks the opportunity to look at different sources of eDNA that animals shed in the environment,” he said. “You don’t have to now have a pure sample, like a biopsy of the animal.”
A visit to Kibale National Park in Uganda gave Gogarten and some of his colleagues a chance to work on an eDNA project that yielded some surprises. Gogarten, along with fellow researcher Christina Lynggaard and other colleagues, swiped low-hanging plant leaves in the park’s tropical forest to see if airborne eDNA had settled on them. A lab analysis later detected dozens of species — mammals like the African elephant and birds like the grey crowned crane.
“It really blew our minds at how much diversity we were detecting just by one swab,” said Gogarten, who also is affiliated with the Department of Applied Zoology and Nature Conservation at the University of Greifswald.

A collection swab after being used to sample a leaf in Kibale National Park, Uganda. Jan Gogarten, Christina Lynggaard, and their colleagues sampled the airborne eDNA that had collected on low-hanging plant leaves.
Visual: Christina Lynggaard
The most unexpected detection was a fish. “We detected a catfish genus, and they live in the rivers there, so they’re around, but they’re not the sort of thing that you would expect would be, like defecating or coming in contact with the leaves,” he said.
Gogarten and his colleagues were perplexed. “Catfish are unique in that they can leave the water, so they sometimes do go on land,” he said. “But our best guess is actually that they probably were eaten by one of the bird species, and the bird was up on a tree and defecated on the leaves.”
As the field expands, the high risk of contamination in the lab remains a struggle for researchers, which Gogarten attributed to the sensitive methods used in eDNA analysis. Other molecules that “may be floating around in lab spaces from previous experiments can cause problems — as can small traces of animal DNA if the extraction areas are used for extracting tissues,” wrote Gogarten in an email to Undark.
To reduce the risk, researchers can set up lab controls and separate workspaces for eDNA analysis, he said. “It’s something that one really needs to be careful with when one is doing eDNA, and it’s something that the field is trying to create, these norms or guidelines that if you want to do eDNA, you have to follow steps to avoid contamination,” said Gogarten. “If we start doing poor and shoddy lab work and publishing it, if we muddy the waters, then we undercut the confidence in the results.”
As promising as eDNA lab methods are, they also have limits. For example, every organism has a sequence, or barcode, associated with it; scientists identify the specimens by comparing barcodes with those archived at DNA reference libraries. But if a species is not listed in the database, then eDNA cannot identify it.
Still, the technology is a boost for assessing biodiversity — meaning taking stock of the variety of species living in certain areas. “Now we can start thinking about projects where you can look at biodiversity along this park edge of Kibale and collect tens of thousands of swabs and do a very high-resolution mapping of biodiversity along these park edges,” he said.
And eDNA could help determine the distribution of biodiversity and help create effective, large-scale management strategies for preserving it, Gogarten said. “It’s an exciting time because the technology is becoming more and more available.”
So far, much of the eDNA research has focused on identifying species in rivers, oceans, lakes, and spring-fed pools like those in southern Arizona national parks. Hubbard said the relative ease and low cost of collecting eDNA samples means the technology is here to stay. He and his team started collecting eDNA in the spring of 2022 and will continue to do so in the coming year.
He expects the technology to keep improving as it provides information that can help natural resource managers to make decisions. In fact, several companies are developing portable lab units to do real-time detections in the field. Those devices are likely several years away from being ready to deploy, and some element of lab work may always be required, he says, but it’s one of the exciting developments for this technology.
“It really blew our minds at how much diversity we were detecting just by one swab.”
For now, he and his team will use eDNA results to come up with a plan for the best way forward. “Our ultimate goal is to have these be healthy wetlands that are sustaining native species again,” he said. “And so we’re trying to take a little pause and get that bigger picture and use the data that we have to inform that.”
Sepulveda expects the national strategy, which he is now working on with the Biden administration, could improve eDNA application and data-management tools in natural resource management, among other possible outcomes. That strategy is scheduled to be made public in June 2024.
“This eDNA strategy is hoping to create a framework, or at least a roadmap, for how to be more coordinated and be more effective and how to take eDNA science, how to continue to improve it for the next 10 to 20 years,” he said.
Meanwhile, the technology keeps gaining a foothold in worldwide conservation. In South Africa, scientists rediscovered a golden mole that had not been sighted since 1937 by tracking its eDNA in sand. In Brazil, eDNA helped scientists rediscover a frog they believed had gone extinct since it had not been seen since 1968. And in the Mediterranean Sea, scientists collected genetic material shed by the endangered and evasive angel shark.


But the vast oceans remain largely unexplored and eDNA is an important tool to unravel the unknown, said Kelly Goodwin, a marine molecular microbiologist with the National Oceanic and Atmospheric Administration. The technology is enhancing the agency’s numerous tools that allow scientists to explore ocean life and protect it, she said.
“There are as many microbes in the ocean as there are stars in the sky,” she said. “There’s a great deal that we do not know about the biology on our planet, including the invisible majority, which is the microbial Earth.”
Goodwin said eDNA technology advances will help answer questions not only about invisible organisms in the deep ocean, but also inform the composition of species — rare, endangered, or invasive — throughout the ecosystem.
“There is a lot of life on this planet that we know very little about,” she said. “And the only lens we have to view it is through their DNA.”