Revealing the Secret Lives of Cells With Advanced Microscopy
Open any biology textbook, and you’ll encounter an artistic rendering of a perfectly round cell, says biophysicist Winfried Wiegraebe. Yet the truth is more complex. Wiegraebe’s team at the Allen Institute for Cell Science in Seattle has been modeling the behavior of individual cells in three dimensions. Among their recent observations: Even with cells of the same type, no two are shaped alike, let alone truly round. “We were surprised,” says Wiegraebe.
More than 350 years after they were first discovered, cells remain in many ways a mystery. How do they differentiate into brain, muscle, or any of the other approximately 200 different human cell types? How do they change as they age? Even more fundamentally, how do these little bags of water and chemicals turn a set of DNA instructions into a dynamic living creature capable of autonomous and coordinated behaviors?
Over the past decade, answers to questions like these are finally coming into focus, thanks to advances in microscopy that have allowed microbiologists to observe living cells interacting, and to peer inside cells at resolutions believed physically impossible for hundreds of years.
It’s not just that someone designed a better microscope — though that’s a big part of it. It’s that biologists have grown multi-disciplinary, recruiting the expertise of other fields to explore the living world at its smallest scale. These collaborations have opened up a whole new way of seeing life inside the cell. The University of California, Berkeley’s Eric Betzig, whose work in the field garnered him a 2014 Nobel Prize in Chemistry, suggests that this new understanding foretells a looming shift in outlook as dramatic as that from Newton to Einstein. While this remains to be seen, there is no doubt that cells have already proved to be far more dynamic and complex than our idealized textbook renderings suggest.
“Now we have the ability to find things in living cells that we never could before — and I’m talking even 10 years ago,” says Steven Ruzin, a microscopist at UC Berkeley and curator of the school’s Golub Collection, which features more than 160 microscopes from the past 350 years. Ruzin likens the sheer quantity of new data to a tsunami.
Since Galileo Galilei, biologists have used microscopes not unlike those in many high schools, with a light source to illuminate the sample. These are governed by the laws of diffraction, limiting resolution to a blur below 250 nanometers (nm). With these microscopes, you can see red blood cells (7,000 nm) and bacteria (typically 300 to 5,000 nm), as well as some of their individual components. But almost all viruses, as well as proteins and many other structures, remain out of view.
Further, because cells are transparent, they are often killed and then stained to provide better contrast. Thus, in the past, scientists generally conducted themselves like coroners, prodding dead bodies and injecting them with toxins. “That was done for hundreds of years,” says Ruzin.
In 1931, Ernst Ruska and Max Knoll figured out how to replace the photons used to illuminate a sample with electrons creating the first electron microscope. Electrons have a much shorter wavelength than light, and thus enable resolutions lower than 50 picometers, or one-twentieth of a nanometer, roughly the distance between the nucleus and the electron in a hydrogen atom.
Electron microscopes provided insight and superb resolution, but they couldn’t penetrate the surface of a cell without researchers slicing it into thin sections, and so biologists continued to work with dead samples. Then, Ruzin says, “all of a sudden someone said, ‘Holy cow, we shouldn’t be doing this because it’s super artificial.’” It occurred to them that coroners are poorly situated to study life.
While in the 1990s and early 2000s biologists were learning the ins-and-outs of keeping cells alive and capable of being viewed and imaged, a breakthrough in microscopy was brewing. Beginning in 2000, a series of papers announced the arrival of super-resolution microscopy: a set of tools that allow scientists to see inside living cells at resolutions of a few dozen nanometers, one-tenth the best resolution long believed possible.
Three scientists, including Betzig, ultimately shared the 2014 Nobel Prize in Chemistry for creating different variations of fluorescence microscopy, which opened up the ability to see what structures and proteins look like inside a living cell, below the limits of light diffraction.
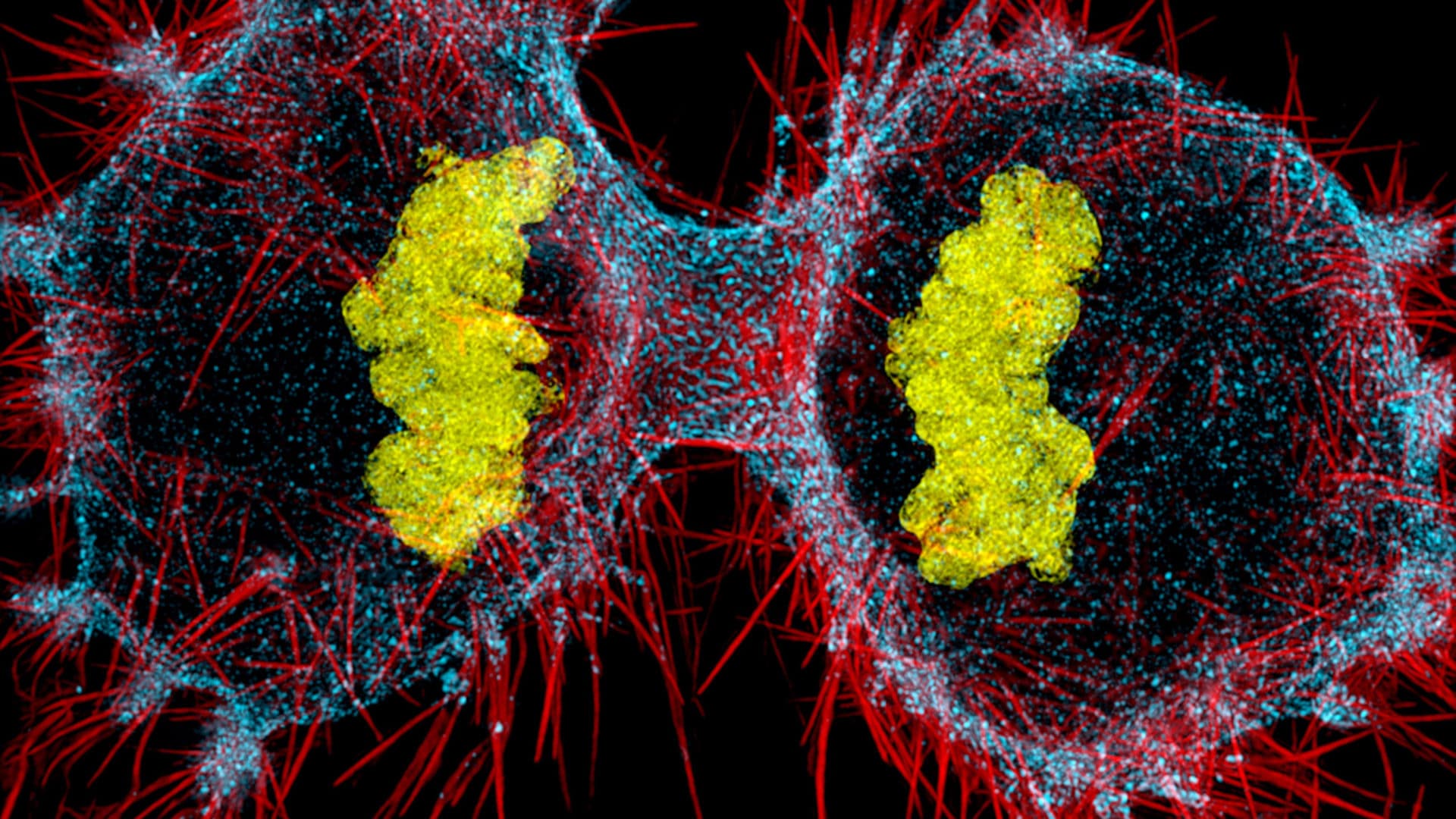
Fluorescence microscopy utilizes fluorophores — molecules that fluoresce when they absorb light of a certain wavelength. These molecules can be genetically encoded, or they can be chemically attached to molecules in the sample, in which case, they are called “probes.” Unlike traditional microscopes, which shroud the sample in light, creating high contrast images, fluorescence microscopy is primarily about darkness, with bits of fluorescence poking through. Imagine a pitch-black club, where you find your friends by their color-coded glow sticks.
Betzig realized that by repeatedly imaging the sample and overlaying the images, his team could reconstruct the sample in 3D. It’s sort of like a strobe light moving and firing quickly about the room to get a 360-degree perspective without upsetting the darkness or the revelers. The advantage of fluorescence microscopy, explains Wiegraebe, is that scientists can label specific structures with fluorophores and directly observe their activity inside the cell.
And yet, even this approach has limitations. Scientists can’t see anything that isn’t color coded. For a variety of reasons — many of them chemical — scientists are limited to a handful of probes per sample, but there are thousands of proteins and dozens of different structures inside human cells, for example. “You won’t find what you’re not looking for,” says Sarah Veatch, a biophysicist at the University of Michigan who uses fluorescence microscopy to study the proteins embedded in cell membranes.
Returning to the nightclub metaphor: You might easily spot your friends with their colorful glowsticks, but you’ll have no idea what’s surrounding them — tables, chairs, other patrons — when those things aren’t similarly illuminated. For a more complete understanding of the cell, Veatch says, researchers are increasingly drawing on tools from other disciplines.
In the last decade, the number of super-resolution approaches has proliferated, many of them enabled by advances in computing and robotics. Some take advantage of high-speed or contrast cameras abetted by the processing power to resolve these images or track small changes. Other approaches, such as cryo-EM, use new flash-freezing technology to effectively (if still fatally) capture all the constituents of the cell. And robotics is also being employed.
“The ability to automate and mechanically tilt the sample and have the sample and beam be stable enough to create 3D images at that resolution [is new],” says Vern Robertson, a veteran sales and product manager at JEOL, a global manufacturer of electron microscopes and other instruments. The sample is then reconstructed from the thousands of images taken at various angles.
“That’s where massive amounts of automation and computer power come in,” says Robertson.
Ben McMorran, a physicist at the University of Oregon, says that with so much information, it is difficult for scientists to identify what is important and new using just their eyes. “Increasingly we have to rely on machine learning and machine vision,” he says. “So we seed our computer with a bunch of data that looks normal and it gets better and better at identifying the stuff that is normal and not normal and then raising that to our attention.”
McMorran is exploring an approach called STEM holography which takes advantage of robotic automation and an electron’s probabilistic travel plans to scan a frozen sample from every possible angle. By picking up the electron’s subtle phase changes he will be able to reconstruct information about the sample’s interior.
It’s an odd irony that as the technology has advanced, in some ways it has outgrown the human eye. While Ruzin still looks through his microscope’s eyepiece, it is only for a moment to line up the sample and flash the fluorescent light. “Then you scooch over to the computer, and you look at the image with your sensitive cameras,” he says.
Sometimes computers can reassemble the data to provide a level of breadth that no single data source can. At the Allen Institute, Wiegraebe and his colleagues are combining real data and predictive modeling to create images of cells. Recently, they unveiled the Integrated Mitotic Stem Cell which demonstrates in interactive 3D how 15 key cell structures change as the cell divides.
“Once you see this process as a complete picture, you can start to uncover new, unexpected relationships and ask and answer completely new questions,” said Rick Horwitz, the executive director of the Allen Institute for Cell Science, in a recent press release.
This has been true for Veatch, who uses super-resolution imaging to study how particular proteins in a cell’s wall or membrane influence how receptors interact on the cell’s surface. This matters for cancer immunotherapies, which train an individual’s immune system to target certain cancers based on a telltale receptor on the cancer cell surface. Veatch was surprised to be able to glean as many insights from what she couldn’t see as what she could in determining the molecular influences around a receptor’s interactions on the cell surface.
She compares these new super-resolution approaches to reading glasses for cell biologists: “It’s not only that you can see small things, but those things are sharper and we’re better able to see the small variations we’re looking for.”
For his part, Betzig believes there’s a lot more going on than we realize, and suggests our understanding up until now has been overly simplistic and perhaps overly mechanistic. In cells as in life, there’s a lot going on that defies easy categorization or explanation. “It’s an environment of tremendous complexity that’s driven by variability and chance,” he says. “It’s going to be a whole new way that people start looking at living systems.”
Chris Parker’s work has appeared in The Hollywood Reporter, Billboard, The Guardian, The Jerusalem Post, NPR, and dozens of alt-weeklies including The Village Voice, LA Weekly, Phoenix New Times, and City Pages.









Comments are automatically closed one year after article publication. Archived comments are below.
A couple of points for clarity:
1. The biology textbooks have not caught up with the extensive work done by scientists, such as those associated with the Human Cell Atlas project, demonstrating that there are far more than a couple hundred different, stable cell types.
2. Contemporary use of the word “microbiologist” generally indicates a scientist who researches microbial organisms (often, but not always, single-celled). While there are certainly microbiologists who study development, using the term doesn’t hit the mark on all those scientists out there working in large multi-organ-system creatures.
Great work unveil the facts of life thanks to all scientist who are doing such amazing work.knowledge about cell will lead to understand natures behaviors
Very surprising research based material. Thanks for researchers and publication.